Parsing¶
Taxonomy Parsing¶
Arelle is a popular open source library for working with XBRL data, and it is used for parsing the taxonomy. Arelle can parse a taxonomy from either a URL to the taxonomy entry point, or a local path to a zipfile containing the entire taxonomy.
The extractor will first take a parsed Taxonomy and construct a taxonomy
object defined in ferc_xbrl_extractor.taxonomy. This is done because the
data structures used by Arelle are not well documented, so they are immediately
translated into custom data structures. After creating these structures, the extractor
will generate a frictionless tabular datapackage,
which contains a schema for the new SQLite DB, as well as useful metadata.
Each table in the SQLite DB is derived from a Link Role in the Taxonomy. This is done by traversing down the Concept tree that is rooted at the Link Role. It will find all Concepts which are leaf nodes of this tree. These leaf nodes are Concepts in the FERC taxonomy that are expected to have Facts reported against them, while Concepts higher in the tree structure exist for defining relationships. The leaf nodes will be sorted by their Period type (either duration or instant), and a table schema will be generated for each of these Period types. The following diagram demonstrates this process.
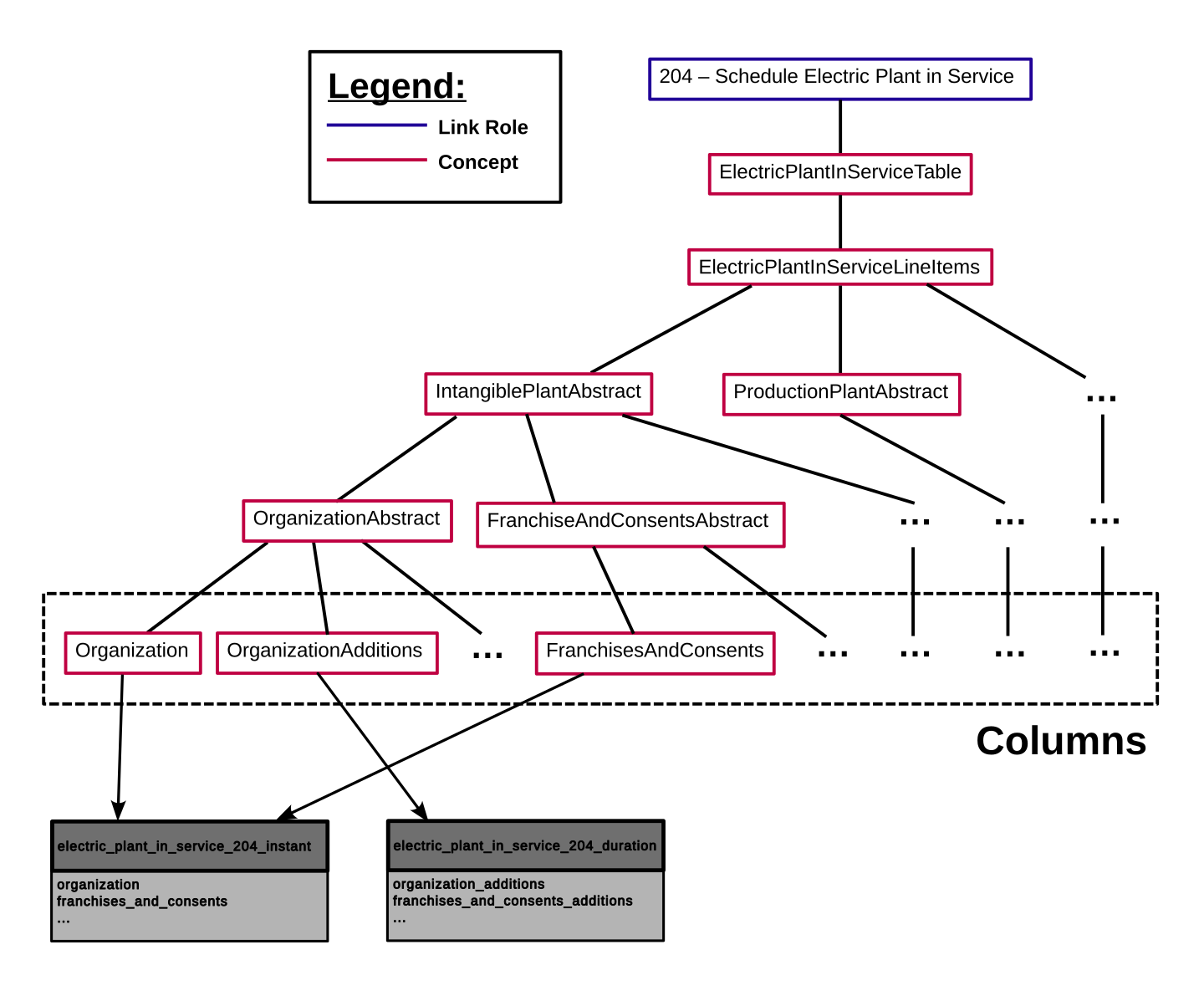
Fig. 1 Example Concept tree from FERC Form 1¶
The Link Role shown here, 204 – Schedule Electric Plant in Service, is turned into
two tables in the resulting database, electric_plant_in_service_204_instant and
electric_plant_in_service_204_duration. The Concepts in the box at the bottom
will become the columns of these tables, and they will be split based on their Period
type.
There will also be columns added to these tables that correspond to the
Context. For duration tables there will always be a start_date,
end_date, and entity_id column. Instant tables will only have a date,
and entity_id columns. There will also be columns added for any Axes
that are defined in the taxonomy. This set of columns created from the Context is
used as the primary key for the table. Using the example from above there are no Axes
defined for the Link Role, 204 – Schedule Electric Plant in Service, so the
primary keys for the two tables will only contain these date and entity columns.
Filing Parsing¶
While Arelle is helpful for parsing taxonomies, it has proven too slow for parsing large sets of Filings. So, we’ve used the lxml library to directly parse filings ourselves. This is not too difficult as this individual XBRL instances are a fairly simple XML structure that is easy to parse. These files contain a list of contexts, and a list of facts that we read into custom data structures.
The first step to parsing a filing is to read all its facts, and save them in a dictionary that is indexed by the Axes in it’s Context. Next, the extractor will loop through the tables created during the taxonomy parsing, and find which facts should end up in each table. This is done by looking up the facts with a Axes that match those in the tables primary key. Each fact that meets this condition, and whose name matches one of the columns in the table will be added to the table. Rows are then created by finding all facts with the same primary key.
Going back to the example above, because there are no Axes defined for the Link Role the extractor would look for all Facts whose Context does not have any Axes. Next, it would it filter that list of Facts to only those whose name matches one of the columns expected for the two output tables. Finally, it would group Facts with identical Contexts (i.e. they have the exact same dates and entity ID’s) into rows, which are added to the table.